NetCoupler: Inferring causal pathways between high-dimensional metabolomics data and external factors
Luke W. Johnston
Clemens Wittenbecher
Fabian Eichelmann
Designed to identify potential causal factors from complex networks
Motivation:
- Moderately high dimensional and complex network data (e.g. metabolomics)
- Derive potential network structure
- Estimate causal pathways:
- From exposure (e.g. exercise) to network
- From network to outcome (e.g. diabetes)
- From exposure to outcome, through the network
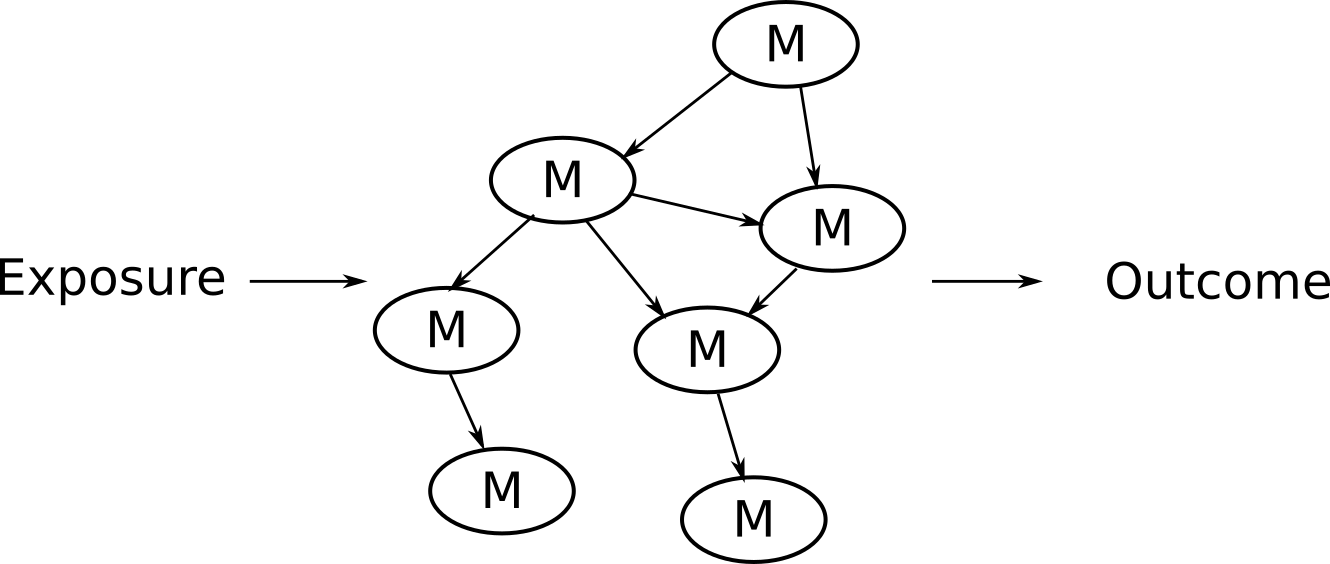
Use NetCoupler to answer questions of this form (M = metabolite).
Hi everyone, I'm going to be talking about NetCoupler, which is an algorithm and R package for inferring causal pathways between high-dimensional metabolomics data and external factors.
We had several motivations for creating NetCoupler, largely because we wanted to use moderately high dimensional and complex network data such as from metabolomics and to be able to answer questions about potential causal pathways that occur through the network. As illustrated by the diagram, we wanted to know how an exposure like exercise might influence a network, how a network might influence an outcome like diabetes, or how an exposure might influence an outcome through the network.
Main features of NetCoupler
Finds most likely network structure
Can include exposure and/or outcome
Identifies potential causal links from, to, and within the network
Main features of NetCoupler
Finds most likely network structure
Can include exposure and/or outcome
Identifies potential causal links from, to, and within the network
Flexible in type of model used (e.g. linear, logistic, cox regression)
Allows adjusting for confounders and covariates
Results are designed to be visualized (e.g. with tidygraph/ggraph packages)
The main features of NetCoupler are that it finds the most likely network structure, it can include exposure and/or outcome variables, and it can identify potential causal links involving the metabolic network.
NetCoupler is also quite flexible in the type of models you can use in it, so you could use models like linear or logistic regression or Cox proportional hazard models. Because these models can be used, you can also adjust for potential confounding factors that might bias the results. Since NetCoupler is based on network graphs, the results are especially designed to be visualized as them too, like with the packages tidygraph or ggraph.
Four basic phases of the algorithm
The NetCoupler algorithm works in four basic phases, illustrated in this diagram.
The first phase is that the structure of the metabolic network is derived using causal structure learning algorithms like the PC-algorithm.
The second phase is where each metabolic variable, called a node, within the network is iteratively selected and set as the index node. Each connected neighbouring nodes are then identified and selected. Here, the index node has three neighbours.
The third phase is where each possible combination of index with neighbouring node is calculated and used in the model. There are three neighbours here, so that would be eight different combinations representing eight models.
The fourth phase is taking all these models and linking them with either an exposure or an outcome variable, as well as any confounding factors. Based on specific thresholds, the link between exposure or outcome and the index node is classified as either direct, ambiguous, or no effect.
Graphical model output allows visual inference of potential pathways
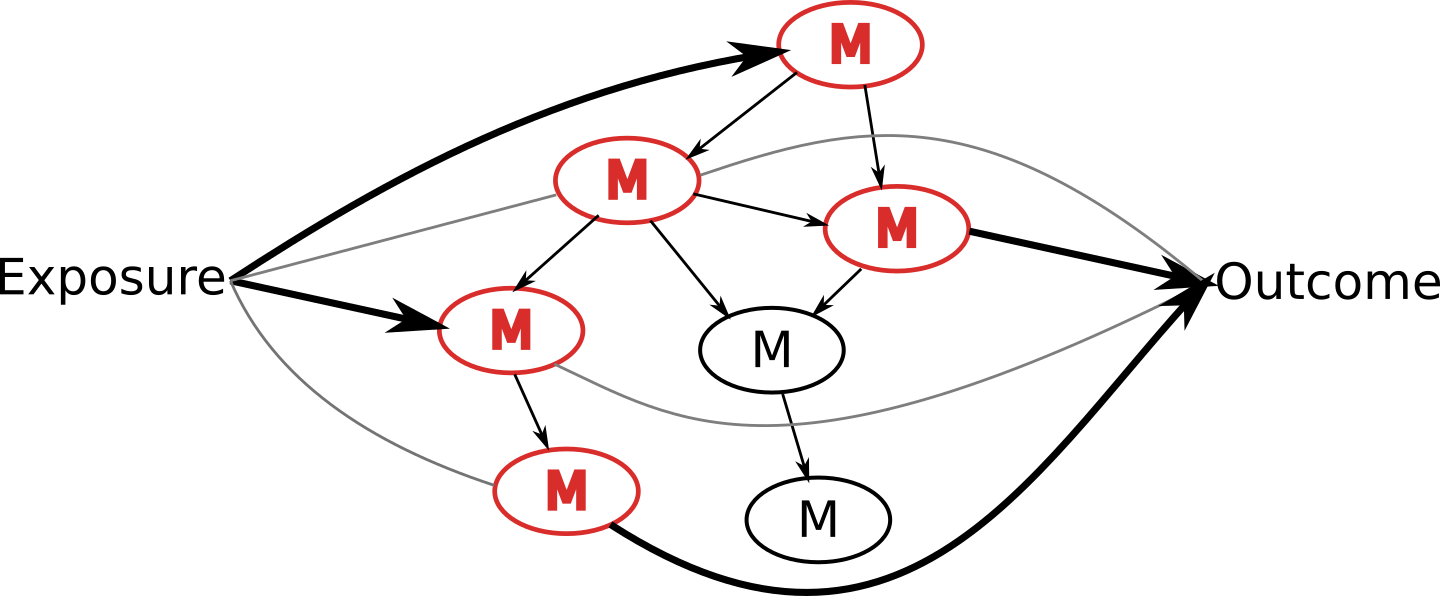
The final graphical model output can allow for visual inference of the potential pathways. For instance, in this example figure, NetCoupler might classify two direct effects, represented by the thicker lines with arrows, and two ambiguous effects, represented by the thinner lines, between an exposure or an outcome and individual metabolic variables. We can then visually trace the pathway from the exposure, through the metabolic variables, and to the outcome, and infer that the metabolic variables along this path, marked as red here, may be along the causal pathway.
Current limitations and areas to improve
- Conceptual:
- Tricky to visualize (too many paths and variables)
- Difficult to interpret output estimates
- Not suited for pure exploration, should have some theoretical basis
- Modeling:
- Heavily relies on p-values
- Only tested on cross-sectional/time-to-event data
Current limitations and areas to improve
- Conceptual:
- Tricky to visualize (too many paths and variables)
- Difficult to interpret output estimates
- Not suited for pure exploration, should have some theoretical basis
- Modeling:
- Heavily relies on p-values
- Only tested on cross-sectional/time-to-event data
- Software:
- Slow performance
- Untested on networks with >25 variables
- Probably not sensible for very high-dimensional data (e.g. genomics)
We're actively working on this R package and there are still limitations and areas to improve.
Conceptually, figuring out how to meaningfully visualize the results has been tricky, because you quickly get too much going on. Because of the pre-processing of the data beforehand, the model estimates can be very difficult to interpret. We also don't believe this algorithm is suited for pure exploration, as there should be at least some theoretical basis for potential causal pathways in your research question.
For modeling, the classification thresholds rely largely on p-values, which can be problematic and we're working on other types of thresholds. We also have only tested NetCoupler on cross-sectional or time-to-event data, so don't know how it would work with other types of data.
Finally, one of the biggest issues is that performance is quite slow. Because of this, we haven't tested it on networks with larger than 25 or so variables and we guess it probably isn't sensible to use on very high dimensional data like genomics.
If you want to see how to use NetCoupler, more detail is on the NetCoupler website found in the footer.
Thanks for listening!
