Reproducibility and code sharing in science
Why it’s important and how to do it
Talk overview
Reproducibility
- Basic principle of scientific method
- Same data, same code, different analyst, same results
- Tools: R Markdown, documentation
Code sharing
- Tied to reproducibility
- For learning and critical review
- Tools: GitHub and Zenodo
- Detailed description
- Includes exactly how analysis was done, ie. based on analysis code
Reproducibility in biomedical science
Already know replication is a major problem
- e.g. Many Labs Project, OSC Project, Reproducibility Project
Don't know extent of reproducibility
- Few studies share data [1]
- Almost no study provides code [2]
Reproducibility in biomedical science
Already know replication is a major problem
- e.g. Many Labs Project, OSC Project, Reproducibility Project
Don't know extent of reproducibility
- Few studies share data [1]
- Almost no study provides code [2]

OSC project: Open Science Collaboration Project
Except maybe bioinformatics, where about 60% of studies do.
Why is it important? 🧐
- Simplest: It's a key pillar of scientific method.
- With modern technology, easy to implement (relative to past)
Why is it important? 🧐
- Simplest: It's a key pillar of scientific method.
- With modern technology, easy to implement (relative to past)
- In biomedical research, poorly implemented or not done at all
- Selected articles: Goldacre, 2019; Munafó, 2017; TOP guidelines; Transparency Checklist; Peng, 2011
(about implementation) I have no training or education (PhD in Nutrition, BSc in Kinesiology) in these and I was able to learn. Though I am a bit obsessive about learning these things so...
There are lots of reasons for this, likely due to:
- Lack of awareness and training
- Difficulty of adoption
- No incentive or reward
- Little to no culture to do it
Keep in mind: Reproducibility is a spectrum
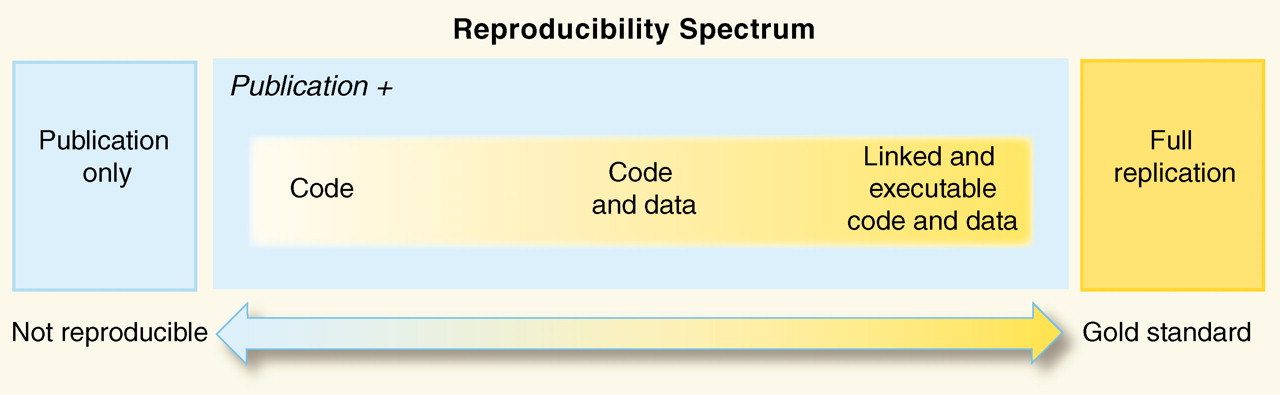
- Should say "Full reproducibility".
Practical ways of doing reproducibility
- Generic ways:
- Documenting scripts and their order, keep them together (in same folder)
- Reproducible document systems
- Pipeline management
- Can't share data? Make fake dataset of the original one.
Practical ways of doing reproducibility
- Generic ways:
- Documenting scripts and their order, keep them together (in same folder)
- Reproducible document systems
- Pipeline management
- Can't share data? Make fake dataset of the original one.
- specific (easy to hard):
- Documenting R scripts, ordering them, and using R Projects
- R Markdown documents
- Pipeline tools (📦: drake, targets)
Why is it important? 🧐
- Tightly tied to reproducibility
- Need code to reproduce results
Why is it important? 🧐
- Tightly tied to reproducibility
- Need code to reproduce results
- Critically review a study's exact analysis
Why is it important? 🧐
- Tightly tied to reproducibility
- Need code to reproduce results
Critically review a study's exact analysis
Builds common standards and best practices
Why is it important? 🧐
- Tightly tied to reproducibility
- Need code to reproduce results
Critically review a study's exact analysis
Builds common standards and best practices
Read others code to learn how to write better
- Code is written to be read by yourself and others [1]
[1]: Otherwise we'd all write in Assembly.
- Reproducibility:
- Code is the exact steps done to data to get results
- Transparent and clear
- Easy to access to any researcher
- Inspectable: (linked to accessibility, but also common language, simple to read, logical, well-reasoned)
- standards: can't do that with hidden code
- how do we get better at writing? By first reading. To get better at coding we need to read others code to know how to write. Like writing text, writing code is done for a reader. If it was purely for the computer, we'd all be writing in Assembly or binary (lowest level programming language)


