MELD Project: Metabolic consequences of Early Life adversity and risk for type 2 Diabetes
Luke W. Johnston
Purpose:
Seek feedback
Show my project
Share about:
- Early life influences
- Analytic technique
- Open science practices
MELD Project Description
Background and rationale
Early life shown to influence risk of diabetes
How do metabolic processes mediate this link (in humans)? 🤷
Difficult to study due to long timespans
Background and rationale
Early life shown to influence risk of diabetes
How do metabolic processes mediate this link (in humans)? 🤷
Difficult to study due to long timespans
Easier now with:
- Registers
- Cohort with registers
- Powerful mediation methods
Some potential mechanisms
Impairs organ development (e.g cell numbers)
Affects insulin sensitivity pathways (e.g. in muscle)
Altered lifestyle behaviours
Excessive stress responses
Public health and clinical relevance
Adversity of trauma:
Refugees
Immigration
Childhood abuse
Poor or unsafe neighbourhood
Public health and clinical relevance
Adversity of trauma:
Refugees
Immigration
Childhood abuse
Poor or unsafe neighbourhood
Adversity of excess:
Childhood obesity
Low-nutrient dense foods
Lack of exercise
For my project, I won't be covering the adversity of excess.
Research questions:
- Overall aim: Better quantify and understand impact of general early life (mainly childhood) conditions on adult metabolic capacity and risk for T2D.
Research questions:
Overall aim: Better quantify and understand impact of general early life (mainly childhood) conditions on adult metabolic capacity and risk for T2D.
Specific objectives:
- How specific conditions affect risk of T2D
- Find mediating pathways of metabolic capacity between adversity and T2D
Research questions:
Overall aim: Better quantify and understand impact of general early life (mainly childhood) conditions on adult metabolic capacity and risk for T2D.
Specific objectives:
- How specific conditions affect risk of T2D
- Find mediating pathways of metabolic capacity between adversity and T2D
- To address 2., need to develop an analytic algorithm
I'm being intentionally vague about meaning of "early childhood conditions" because what conditions I use depends on the data that's available and what may better represent "adversity". This will require some exploratory work at objective 1. to then use in objective 2.
NetCoupler, which does direct and mediating pathway estimation
Current Analytic Problem
Modern studies generate lots of metabolic data
-omics type data
Metabolic biomarkers
High dimensionality
Complex networks
"Traditional" analysis may use: Dimensionality reduction
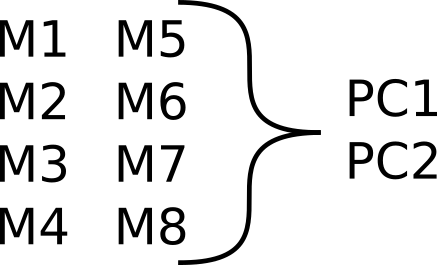
Reducing number of variables with PCA
This has the advantage of making things simpler while trying to maximize variance in the data. Afterward you can do modelling on each principal component. The disadvantage of this approach is that it loses a lot of information since the interdependence and connections between variables it not maintained.
"Traditional" analysis may use: Many regression-type models
O1 = M1 + covariatesO1 = M2 + covariates...O1 = M7 + covariatesO1 = M8 + covariatesSome ways you might go about analyzing this data is by running many regression models, one for each metabolic variable for instance.
This of course has problems since you're simply running a bunch of models and not taking account of the inherent interdependencies between variables.
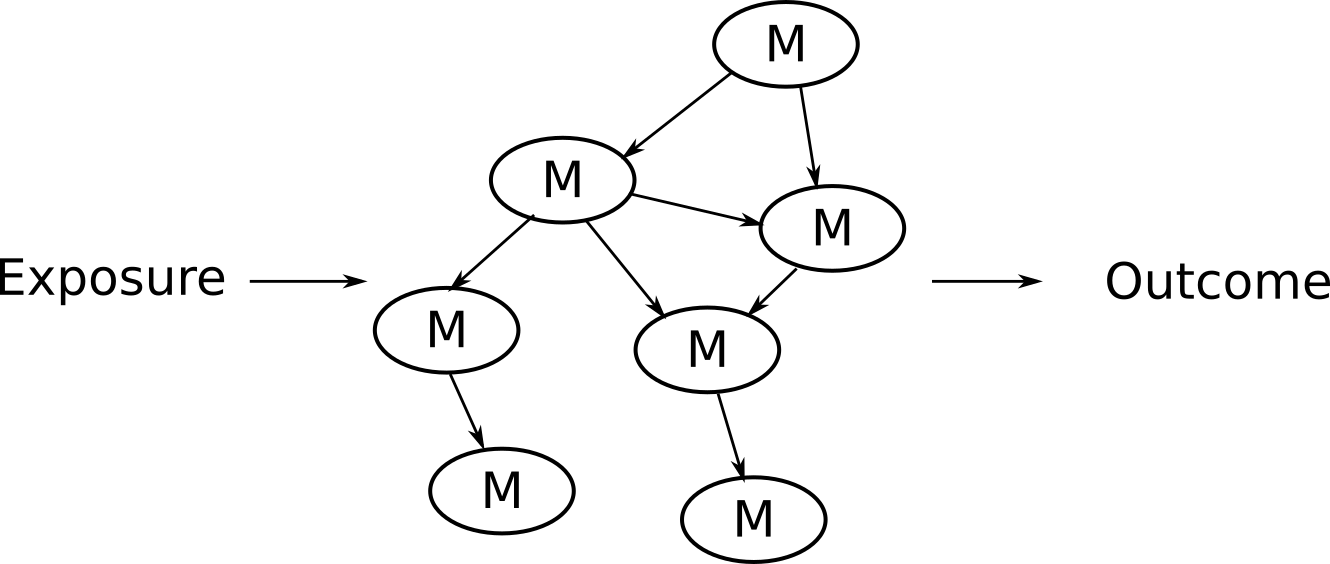
This approach is nice in that you can extract information about the connection between metabolic variables. But there is no way to incorporate the disease outcome with this approach and in order to construct the network properly most methods require you provide a prespecified base network, which you might not know.
But, what if we...
want info about network structure?
don't know the network structure?
But, what if we...
want info about network structure?
don't know the network structure?
have an exposure, metabolites, and outcome?
But, what if we...
want info about network structure?
don't know the network structure?
have an exposure, metabolites, and outcome?
are interested in causal links?
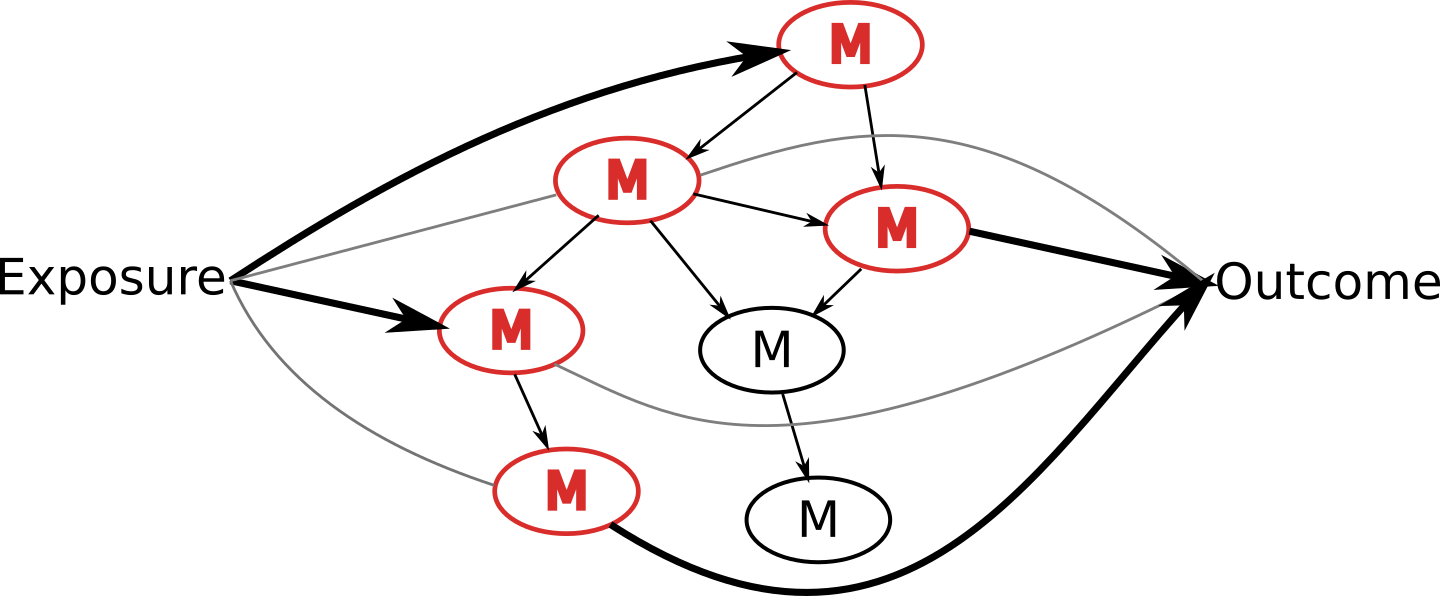
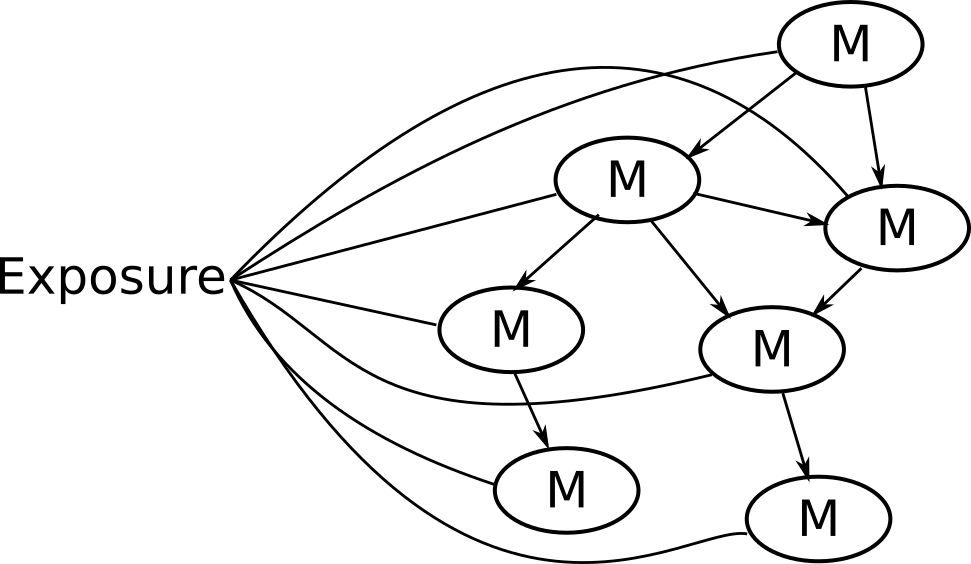
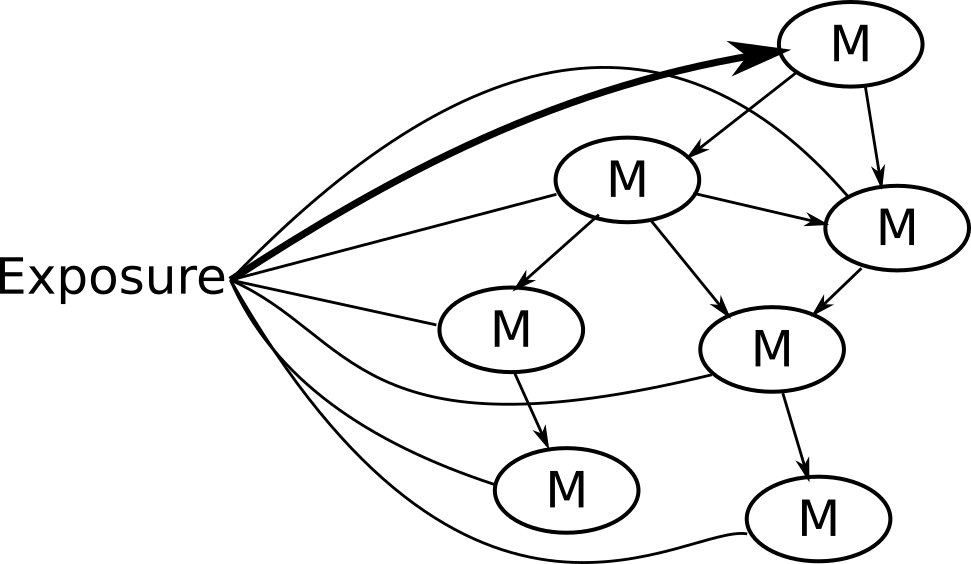
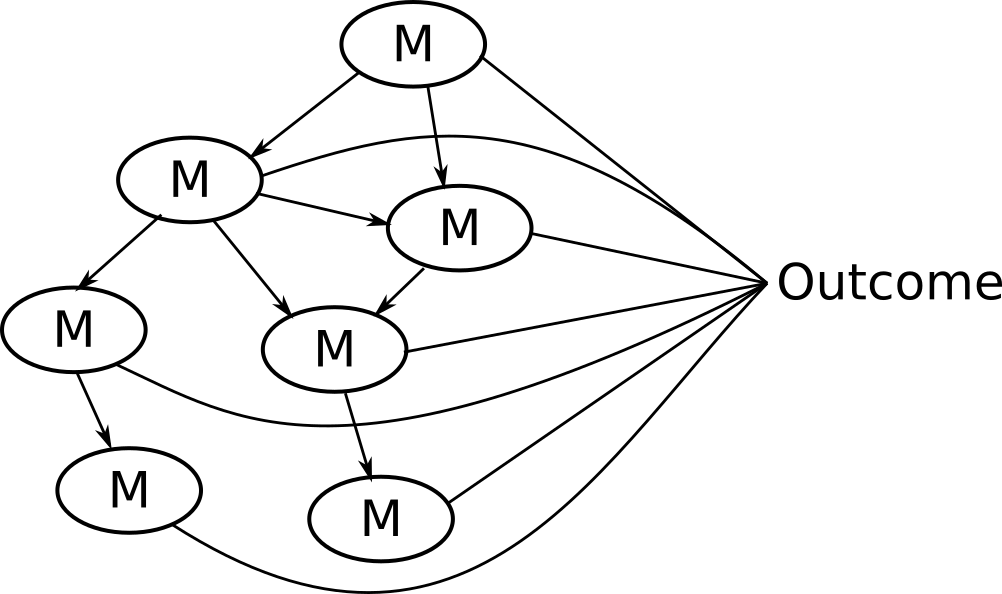
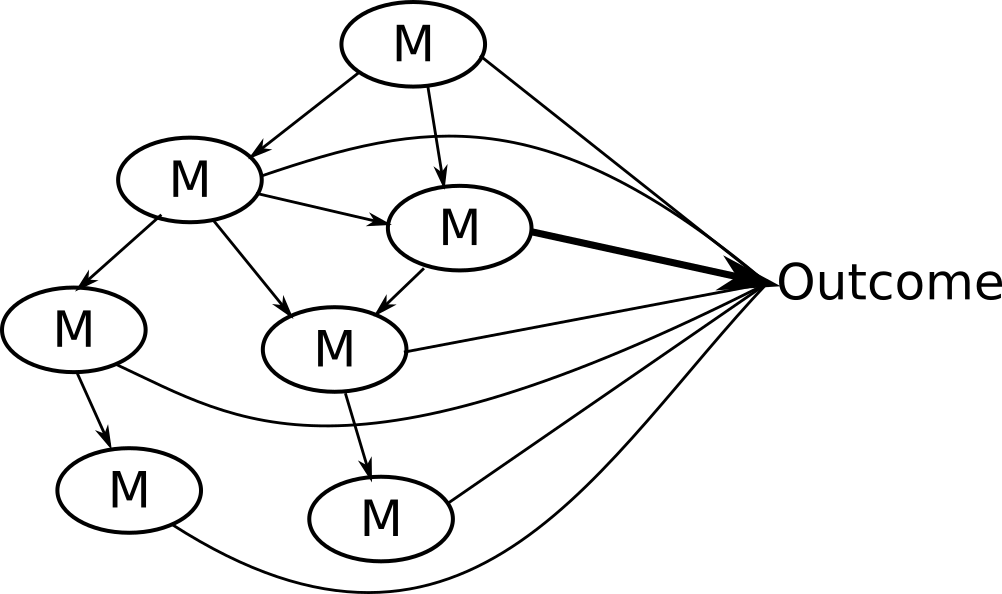
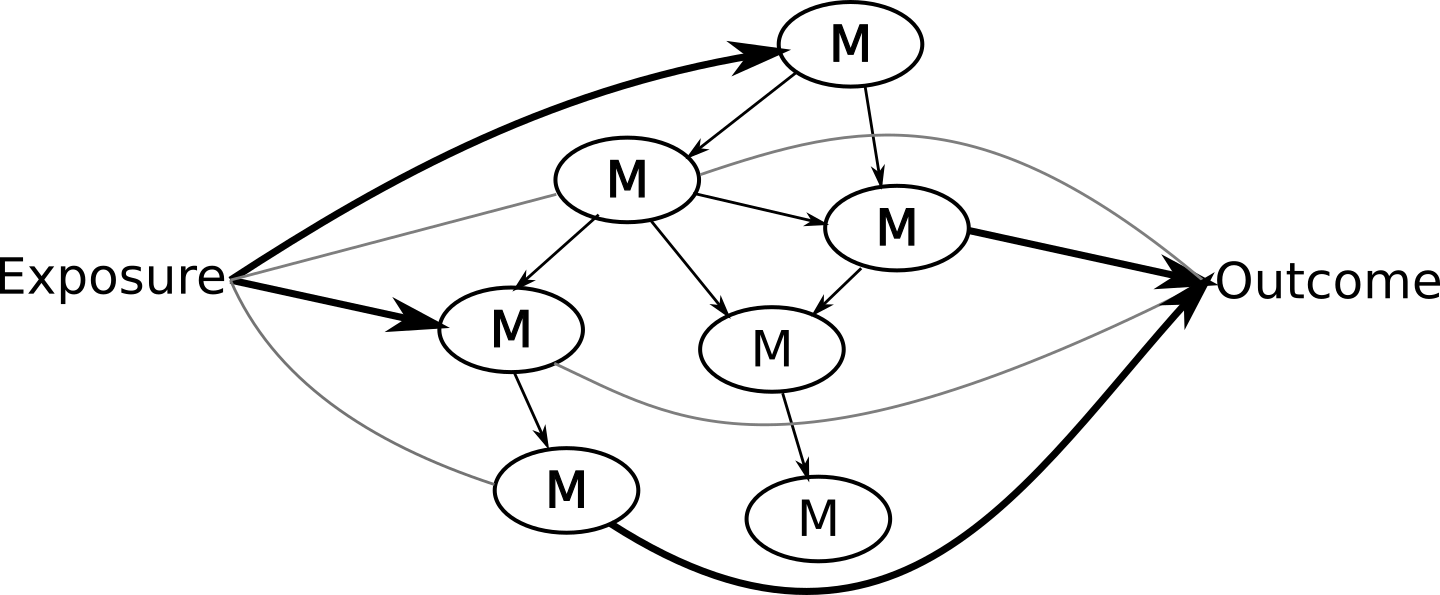
Potential solution: NetCoupler
Causal structure learning that ...
Finds most likely network structure
Allows inclusion of exposure and outcome
Identifies causal links between and within network
NetCoupler algorithm was developed by Clemens Wittenbecher.
Current state of project: Just started
(Trying to) Adhere to open scientific practices:
- Project proposal: lwjohnst.gitlab.io/dda-pdf
Current state of project: Just started
(Trying to) Adhere to open scientific practices:
Project proposal: lwjohnst.gitlab.io/dda-pdf
Developing NetCoupler: 📦 github.com/NetCoupler
Current state of project: Just started
(Trying to) Adhere to open scientific practices:
Project proposal: lwjohnst.gitlab.io/dda-pdf
Developing NetCoupler: 📦 github.com/NetCoupler
Developing protocol to register study: lwjohnst.gitlab.io/meld-protocol
About transparency of process to making claim.
Question:
How many know or have heard or know about open science?
Question:
...or open access, open methods, open data, or open source?
Open science: Terms and meanings
| Term | Meaning |
|---|---|
| Open science | Freely available, openly licensed material for all things related to scientific activity |
| Open access | Free, unrestricted, publicly available published articles |
| Open data | Freely available, re-usable, openly licensed data |
| Open source/code | Freely available, re-usable, openly licensed scientific code used in generating results |
| Open methods/protocol | Freely available, re-usable, openly licensed methods and protocols used to create the data |
Thanks for listening 😁
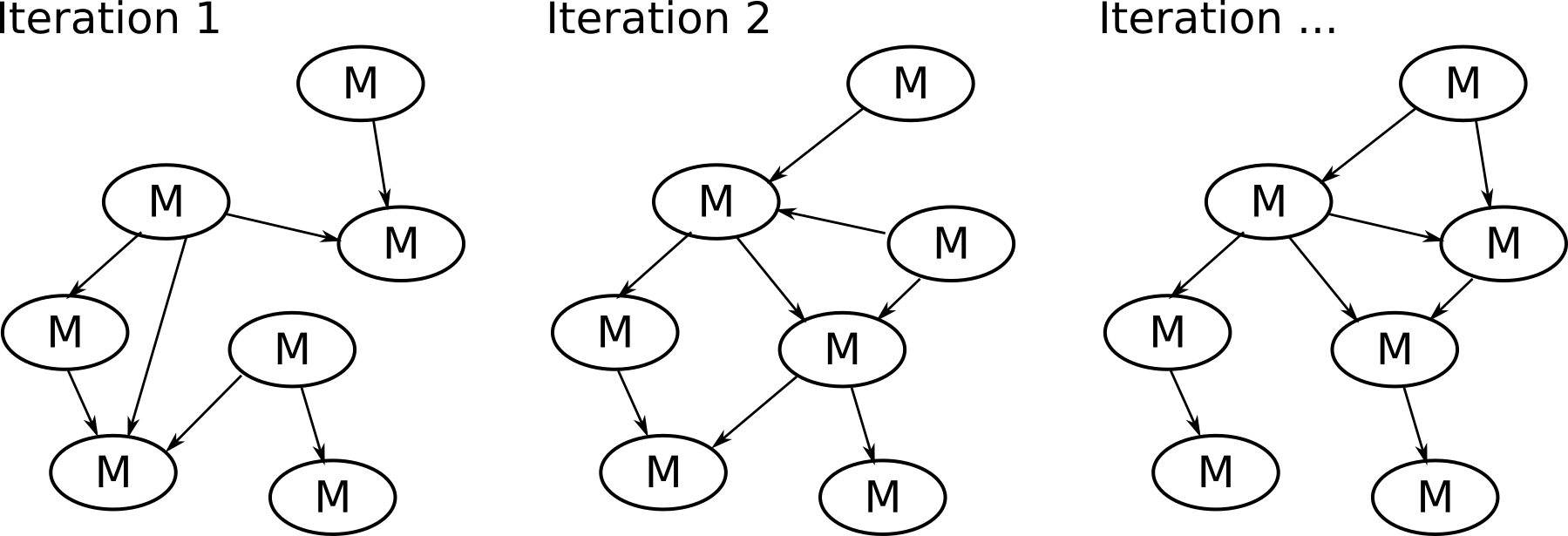
Algorithm (steps) used in NetCoupler
4. Repeat identifying links, find ambiguous, delete indirect links

If you were interested in linkages with an exposure on the network, you could stop here.
7. Repeat identifying links, delete indirect links, find ambiguous
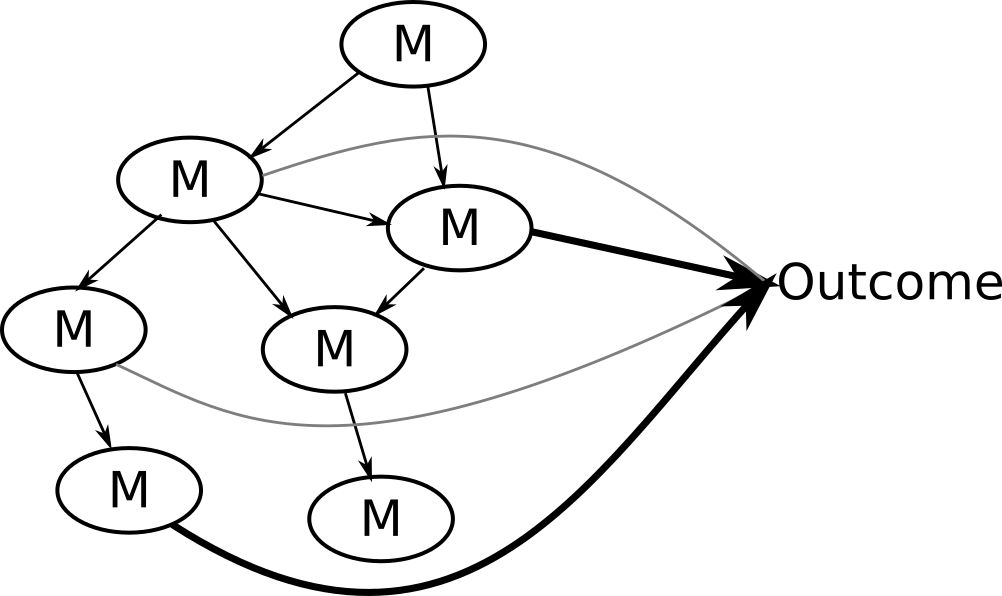
You could stop here if you were interested in linkages with the network and an outcome.