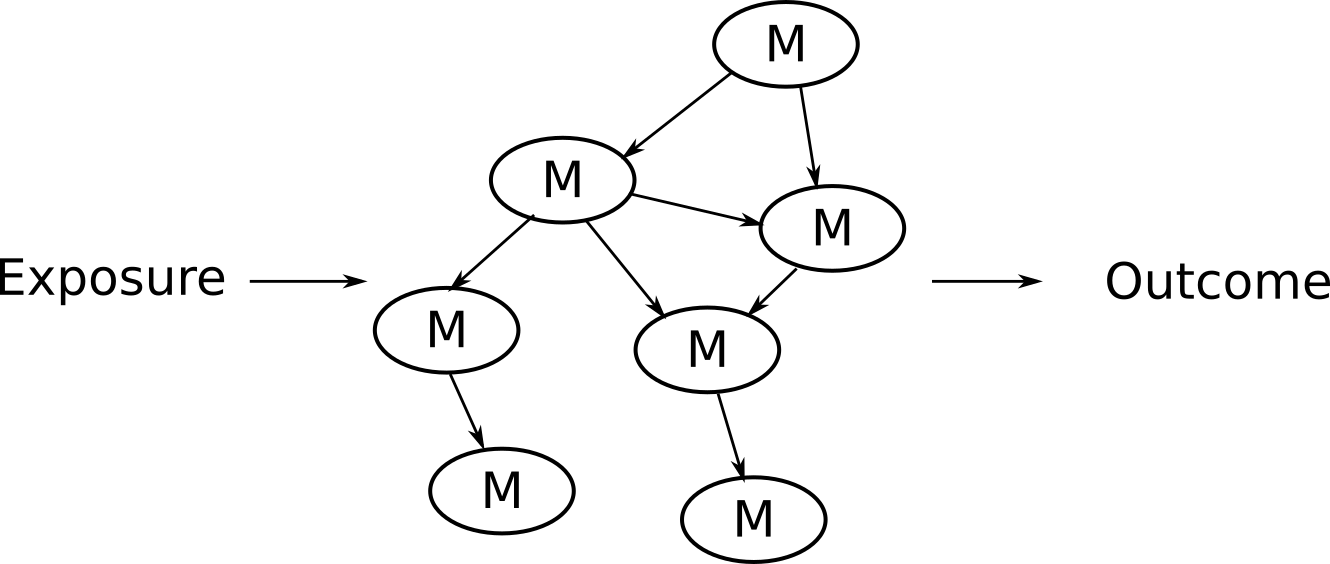
NetCoupler: Inferring causal pathways between high-dimensional metabolic data and external factors
Luke W. Johnston
Steno Diabetes Center Aarhus, Denmark
Outline:
History and background on the analytic problem
NetCoupler implementation and development
Examples using NetCoupler
Current challenges
Amount of data generated is massive
-omics type data
Metabolic biomarkers
High dimensionality
Complex networks
Want to ask questions using this type of data, some way to handle that complexity.
Potential analysis: Dimensionality reduction
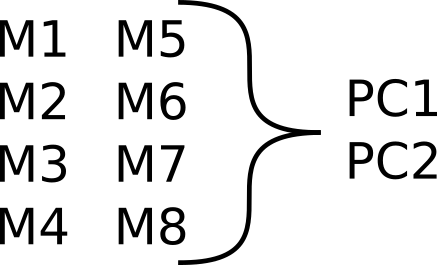
Reducing number of variables with PCA.
This has the advantage of making things simpler while trying to maximize variance in the data. Afterward you can do modelling on each principal component. The disadvantage of this approach is that it loses a lot of information since the interdependence and connections between variables it not maintained.
Potential analysis: Many regression-type models
O1 = M1 + covariatesO1 = M2 + covariates...O1 = M7 + covariatesO1 = M8 + covariatesSome ways you might go about analyzing this data is by running many regression models, one for each metabolic variable for instance.
This of course has problems since you're simply running a bunch of models and not taking account of the inherent interdependencies between variables.
This approach is nice in that you can extract information about the connection between metabolic variables. But there is no way to incorporate the disease outcome with this approach and in order to construct the network properly most methods require you provide a prespecified base network, which you might not know.
But, what if...
want info about network structure?
don't know the network structure?
But, what if...
want info about network structure?
don't know the network structure?
have an exposure, metabolites, and outcome?
Initial development
Developed by Clemens Wittenbecher for his PhD thesis
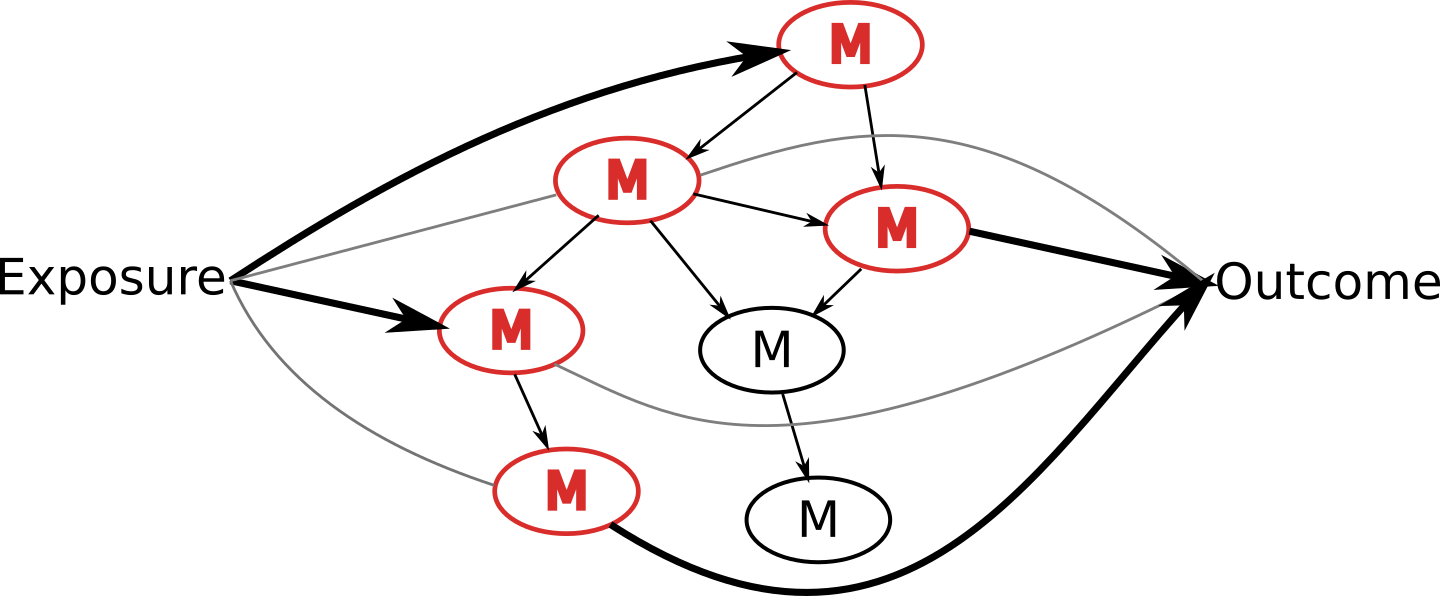
Algorithm that:
- Finds most likely network structure
- Allows inclusion of exposure and outcome
- Identifies causal links from, to, and within the network
A key is that the algorithm is flexible enough to handle different types of models
Infer (potentially) causal pathways with graphical model output
Developing NetCoupler as an R package
- github.com/NetCoupler
- Goal: Submit to CRAN by mid-2021.
Met him at EDEG, asked if it was an R package. Started working together after that.
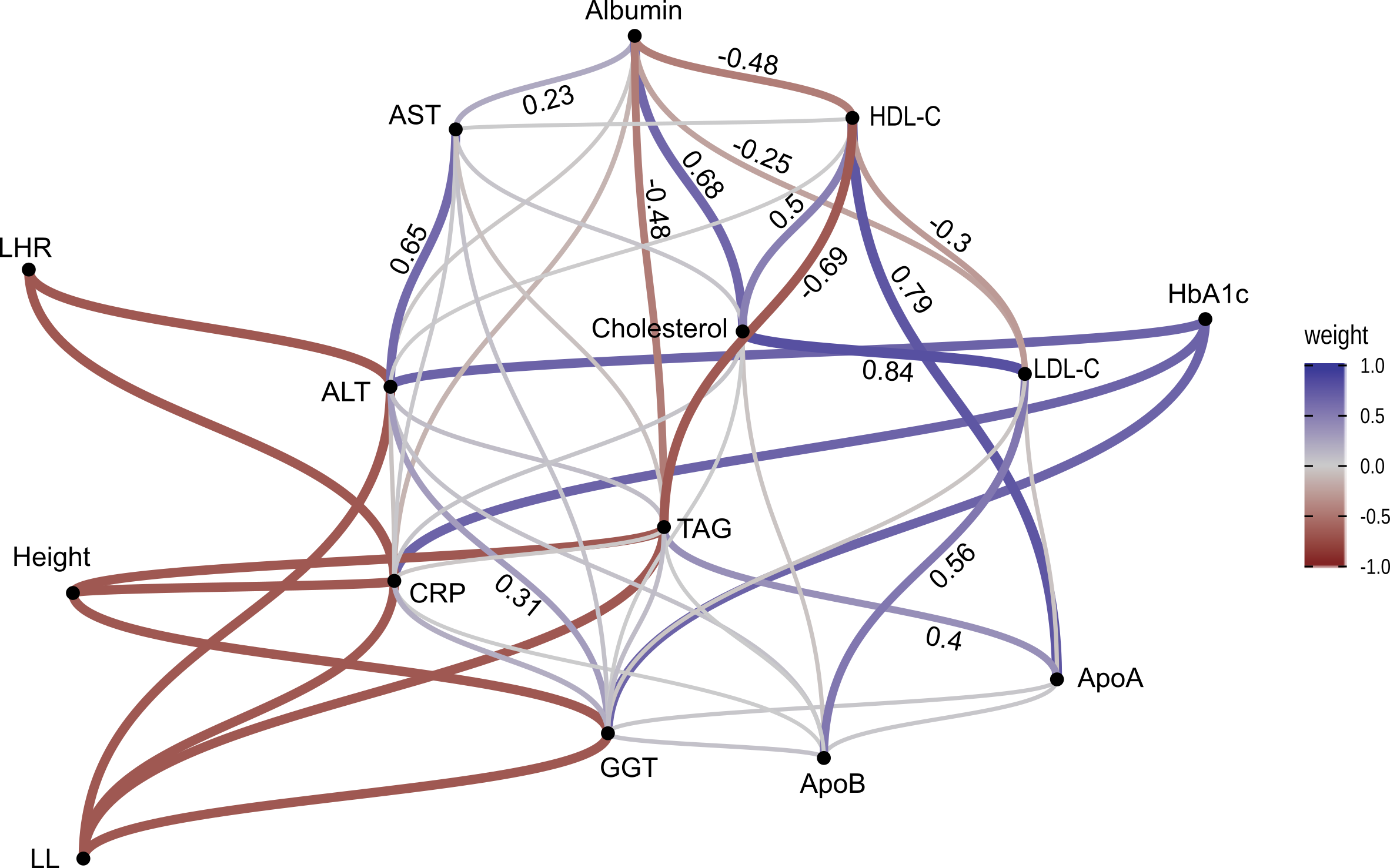
UK Biobank: Metabolic pathways between components of stature and HbA1c
UK Biobank characteristics
- Basics:
- ~480,000 participants
- Cross-sectional
- Stature measures
- Various demographics
- Metabolic variables:
- Cholesterol
- Albumin
- Alanine Aminotransferase
- Apolipoprotein A and B
- Aspartate Aminotransferase
- C-reactive Protein
- Gamma Glutamyltransferase
- HDL and LDL Cholesterol
- Triglycerides
Several limitations or improvements
- Mainly, performance can be slow
- E.g. larger data or networks
Several limitations or improvements
- Mainly, performance can be slow
- E.g. larger data or networks
- Untested on larger networks
Several limitations or improvements
- Mainly, performance can be slow
- E.g. larger data or networks
Untested on larger networks
Untested on non-cross-sectional/time-to-event data
Several limitations or improvements
- Mainly, performance can be slow
- E.g. larger data or networks
Untested on larger networks
Untested on non-cross-sectional/time-to-event data
Visualizing can be tricky
Several limitations or improvements
- Mainly, performance can be slow
- E.g. larger data or networks
Untested on larger networks
Untested on non-cross-sectional/time-to-event data
Visualizing can be tricky
Interpreting estimates can be tricky